research
genomics and evolution of aging, genome structure, and cellular and organismal diversity
The Sudmant Lab at UC Berkeley uses computational, statistical, and experimental methods to interrogate genetic and molecular phenotypic diversity at both the organismal and cellular level. We study the evolution, causes, and consequences of aging as well as the evolution of genome structure and cellular diversity.
The Evolution of Aging
 There is greater than 100,000-fold variation in lifespan among organisms on this planet. We use genome sequencing to identify the genetic underpinnings of differences in longevity between organisms and to understand the evolutionary contexts in which differences in longevity emerge. This work involves employing primary tissue and cell lines from organisms such as the Rockfish to generate high-quality reference genomes for comparative genomics. Additionally, we probe cellular phenotypes such as gene expression and splicing using assays such as RNAseq to complement our genome sequencing efforts and to gain insights into the molecular causes of differences in longevity.
There is greater than 100,000-fold variation in lifespan among organisms on this planet. We use genome sequencing to identify the genetic underpinnings of differences in longevity between organisms and to understand the evolutionary contexts in which differences in longevity emerge. This work involves employing primary tissue and cell lines from organisms such as the Rockfish to generate high-quality reference genomes for comparative genomics. Additionally, we probe cellular phenotypes such as gene expression and splicing using assays such as RNAseq to complement our genome sequencing efforts and to gain insights into the molecular causes of differences in longevity.
The Evolution of Genome Structure and Diversity
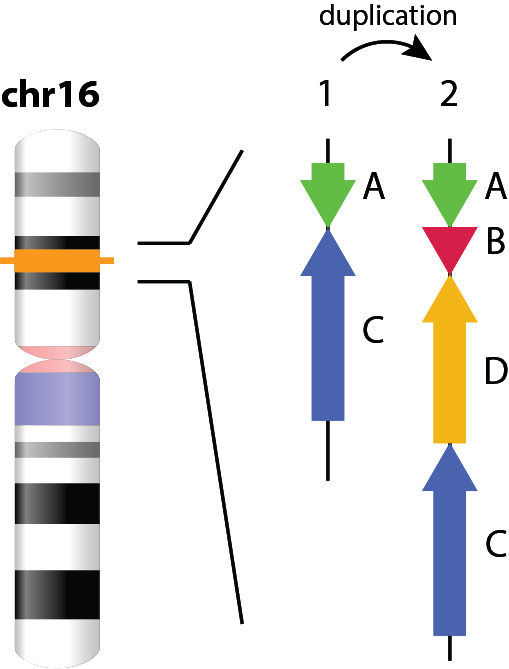 The genome is the source of all phenotypic variability, however, much of the remarkable complexity of genome architectures have been intractable to most sequencing and assembly techniques. Even well studied genomes such as the human genome remain incomplete in some of the most challenging repetitive regions. Novel technologies however have recently emerged that allow such regions to be interrogated and explored across the diversity of life. Our lab employs novel sequencing, mapping, and computational techniques to understand how vertebrate genome structures have evolved in the context of different evolutionary histories, selective pressures and life history strategies.
The genome is the source of all phenotypic variability, however, much of the remarkable complexity of genome architectures have been intractable to most sequencing and assembly techniques. Even well studied genomes such as the human genome remain incomplete in some of the most challenging repetitive regions. Novel technologies however have recently emerged that allow such regions to be interrogated and explored across the diversity of life. Our lab employs novel sequencing, mapping, and computational techniques to understand how vertebrate genome structures have evolved in the context of different evolutionary histories, selective pressures and life history strategies.
Aging and Molecular Fidelity: gene regulation and mutation
 As organisms age many key molecular processes become impaired, however, the full extent of this phenomenon and how it is impacted by external factors such as cell type, genotype, and environment is not well understood. We study how the fidelity of molecular processes is impacted by age and stress in different cellular, organismal, environmental, and genetic contexts. To assay how molecular fidelity is impacted by age and stress we employ high-throughput molecular assays such as RNAseq, single cell sequencing, and ribosome profiling in both primary tissues and cell culture. We also analyze sequence data to identify somatic mutations representing historical errors in the fidelity of DNA replication and repair. We analyze these data in combination with publicly available massive scale datasets to gain insights into the molecular etiology of cellular aging and stress response.
As organisms age many key molecular processes become impaired, however, the full extent of this phenomenon and how it is impacted by external factors such as cell type, genotype, and environment is not well understood. We study how the fidelity of molecular processes is impacted by age and stress in different cellular, organismal, environmental, and genetic contexts. To assay how molecular fidelity is impacted by age and stress we employ high-throughput molecular assays such as RNAseq, single cell sequencing, and ribosome profiling in both primary tissues and cell culture. We also analyze sequence data to identify somatic mutations representing historical errors in the fidelity of DNA replication and repair. We analyze these data in combination with publicly available massive scale datasets to gain insights into the molecular etiology of cellular aging and stress response.
The Evolution of Cellular Diversity
 Phenotypic differences between species are driven by changes in gene expression and gene regulatory programs. However, even across hundreds of millions of years many key morphological features and gene expression signatures in different organisms are conserved. We are interested in the comparative evolution of molecular and cellular diversity in different organs and how within species diversity compares to between species diversity. These studies involve single cell RNAseq on different tissues of diverse species and developing comparative genomic strategies for unique molecular processes.
Phenotypic differences between species are driven by changes in gene expression and gene regulatory programs. However, even across hundreds of millions of years many key morphological features and gene expression signatures in different organisms are conserved. We are interested in the comparative evolution of molecular and cellular diversity in different organs and how within species diversity compares to between species diversity. These studies involve single cell RNAseq on different tissues of diverse species and developing comparative genomic strategies for unique molecular processes.